We would like to invite to a Crash Course on NIFTy (Numerical Information Field TheorY) and on the new GRAVITY imaging tool. The course will take place from August 24th to 27th, 2021 in an hybrid in-person/video-conference format.
The first half of the course will be done by Philipp Arras (MPA/TUM) on the Python package NIFTy. NIFTy is a versatile library that can be used in a variety of inference and data analysis applications. Examples include (x-ray, optical, radio, VLBI) imaging, data calibration applications and the reconstruction of light curves. NIFTy excels in high-dimensional Bayesian inference applications. Basic information can be found in the NIFTy documentation.
The second half of the course will be done by Julia and is specific to GRAVITY imaging with the help of NIFTy.
Closed.
All times refer to CEST.
The course includes hands-on sessions during which the participants can experiment with NIFTy themselves. For this, please install NIFTy beforehand. The latest stable version is available on PyPI and can be installed via:
pip3 install nifty7In order to check that the installation was successful, you may run the following script.
import nifty7 as ift
import numpy as np
dom = ift.RGSpace([500, 500], harmonic=True)
HT = ift.HarmonicTransformOperator(dom)
pdom = ift.PowerSpace(dom)
PD = ift.PowerDistributor(dom, pdom)
S = ift.DiagonalOperator(PD(ift.PS_field(pdom, lambda k: 100./(20. + k**3))))
s = S.draw_sample_with_dtype(dtype=np.float64)
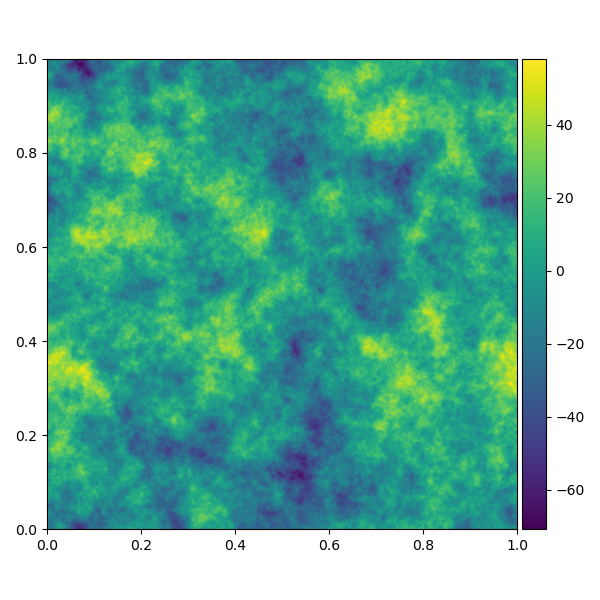
ift.single_plot(HT(s), name="nifty_test.png")If the resulting file nifty_test.png looks like the
following, NIFTy is correctly installed.

Please find the tutorial scripts here.
# This program is free software: you can redistribute it and/or modify
# it under the terms of the GNU General Public License as published by
# the Free Software Foundation, either version 3 of the License, or
# (at your option) any later version.
#
# This program is distributed in the hope that it will be useful,
# but WITHOUT ANY WARRANTY; without even the implied warranty of
# MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the
# GNU General Public License for more details.
#
# You should have received a copy of the GNU General Public License
# along with this program. If not, see <http://www.gnu.org/licenses/>.
#
# Copyright(C) 2013-2021 Max-Planck-Society
#
# NIFTy is being developed at the Max-Planck-Institut fuer Astrophysik.
import numpy as np
import nifty7 as ift
from helpers import generate_wf_data, plot_WF
# Want to implement: m = Dj = (S^{-1} + R^T N^{-1} R)^{-1} R^T N^{-1} d
ift.random.push_sseq_from_seed(111)
position_space = ift.RGSpace(256, 1/256)
data_space = ift.UnstructuredDomain(256)
prior_spectrum = lambda k: 1/(10. + k**2.5)
data, ground_truth = generate_wf_data(position_space, prior_spectrum)
ground_truth = ift.makeField(position_space, ground_truth)
data = ift.makeField(data_space, data)
plot_WF("1_data", ground_truth, data)
harmonic_space = position_space.get_default_codomain()
HT = ift.HarmonicTransformOperator(harmonic_space, position_space) # basically just a Fast Fourier transform
# Define prior covariance in harmonic space
Sh = ift.create_power_operator(harmonic_space, prior_spectrum)
mask = np.ones(256, np.bool)
mask[50:150] = False
mask[180:190] = False
mask = ift.makeField(position_space, mask)
#class BoilerplateLinearOperator(ift.LinearOperator):
# def __init__(self, mask):
# self._domain = ift.makeDomain(...)
# self._target = ift.makeDomain(...)
# self._capability = self.TIMES | self.ADJOINT_TIMES
#
# def apply(self, x, mode):
# self._check_input(x, mode)
# if mode == self.TIMES:
# raise NotImplementedError
# elif mode == self.ADJOINT_TIMES:
# raise NotImplementedError
# raise RuntimeError
class MaskingOperator(ift.LinearOperator):
def __init__(self, mask):
n_data_points = np.sum(mask.val)
assert mask.s_sum() == n_data_points
self._domain = ift.makeDomain(mask.domain)
self._target = ift.makeDomain(ift.UnstructuredDomain(n_data_points))
self._capability = self.TIMES | self.ADJOINT_TIMES
self._mask = mask.val
def apply(self, x, mode):
self._check_input(x, mode)
if mode == self.TIMES:
x = x.val # Unwrapping from field
res = x[self._mask] # Actual code
return ift.makeField(self._target, res) # Wrapping as field
else: # mode == self.ADJOINT_TIMES
x = x.val # Unwrapping from field
res = np.zeros(self._domain.shape, dtype=x.dtype)
res[self._mask] = x
return ift.makeField(self._domain, res) # Wrapping as field
mask_op = MaskingOperator(mask)
ift.extra.check_linear_operator(mask_op)
data_space = mask_op.target
data = ift.makeField(data_space, data.val[mask.val])
R = mask_op @ HT
N = ift.ScalingOperator(data_space, 0.1)
j = (R.adjoint @ N.inverse)(data)
#ift.single_plot(j)
ic = ift.GradientNormController(iteration_limit=256)
metric = ift.WienerFilterCurvature(R, N, Sh, ic, ic)
D = metric.inverse
# Same as
#D = (Sh.inverse + R.adjoint @ N.inverse @ R).inverse
m = D(j)
# plot_WF("1_result", ground_truth, data, HT(m))
n_samples = 10
samples = [HT(m + D.draw_sample()) for _ in range(n_samples)]
ift.single_plot(mask_op.adjoint(ift.full(data_space, 1.)), name="1_data_coverage.png")
ift.single_plot(samples, name="1_posterior_samples.png")
# plot_WF("1_result_sample", ground_truth, data, samples[0])
# plot_WF("1_result_uncertainty", ground_truth, data, HT(m), samples=samples)